Yeast experiment: Yeast Microarray Protocol
Other protocol, Hybridization protocol
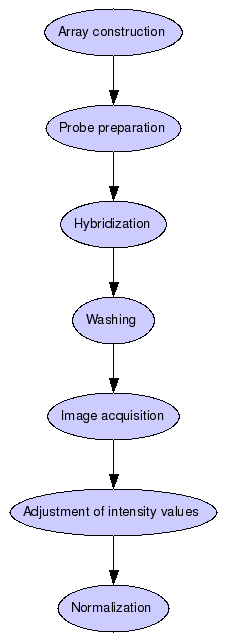
Array construction:
The yeast oligonucleotide array was constructed using the S. cerevisiae Genome Oligo Set ™ (Operon Technologies, CA, USA) composed of 6240 optimized oligonucleotides (70mers) each representing one yeast gene.
Probe preparation and hybridization:
We used the indirect labeling method. Briefly, the reactive amine derivative of dUTP, 5-(3-
aminoallyl)-2?-deoxyuridine 5?-triphosphate (Sigma) was incorporated into cDNA using the Superscript II reverse transcriptase (Invitrogen) and oligo dT (Invitrogen) and random examers (Roche). After synthesis of cDNA (2–3 h at 42 °C), RNA was hydrolyzed by addition of sodium hydroxide and EDTA to a final concentration of 100 mM and 10 mM, respectively and incubated at 65 °C for 10 min. The hydrolysis reaction was neutralized with 1 M HEPES. After removing free nucleotides by purification and concentration using Microcon-30 microconcentrators, the aminoallyl-labeled samples were coupled to succinimidyl ester of cyanine-3 (Cy3) and cyanine-5 (Cy5) (Amersham) combined with 1 M NaHCO3, pH 9. Coupling took place in the dark at 25 °C for 1 h. Appropriate Cy3 and Cy5 labeled cDNA samples were purified following Qiagen Qiaquick PCR Purification Kit instructions. Poly-dA (12–18 mer), 20× SSC, and HEPES pH 7.0 were added. After the resultant mix was filtered through a Millipore 0.45 ?M filter, 10% SDS was added. The samples were incubated for 2 min at 100 °C and cooled in a microcentrifuge prior to loading and then applied to microarray. Incubation took place at 65 °C for 12–15 h. Hybridized slides were washed in a solution of water, 20× SSC, and 10% SDS, rinsed in water and 20× SSC, and dried via centrifugation for 2 min at 1000 rpm.
Image acquisition:
The arrays were scanned immediately. Each comparison was performed in duplicate. Fluorescent cDNA bound to the microarray was detected with a GenePix 4000 microarray scanner (Axon Instruments, Foster City, CA), using the
GenePix 4000 software package to quantify microarray fluorescence. Intensity values were adjusted by subtracting surrounding background from spots. The median of spot intensities was corrected for background. To eliminate signals that are most prone to estimation error, any spot was excluded from analysis if both the Cy3 and Cy5 mean fluorescence signals were within two standard deviations of the mean background signals for that spot. This procedure avoids artificially inflated measurements of expression due to low signals. Additional to eliminating flagged spots, spots were also visually inspected and flawed ones discarded from the analysis. Data were normalized to mean ratio intensity.
We can provide one experiment with 32 micorarrays, two experiments with 8 microarrays and one experiment with 12 micorarrays.
-
Array construction
The yeast oligonucleotide array is constructed using the S. cerevisiae Genome Oligo Set ™ (Operon Technologies, CA, USA) composed of 6240 optimized oligonucleotides (70mers) each representing one yeast gene.
-
Probe preparation
Indirect labeling method. Briefly, the reactive amine derivative of dUTP, 5-(3-
aminoallyl)-2?-deoxyuridine 5?-triphosphate (Sigma) is incorporated into cDNA using the Superscript II reverse transcriptase (Invitrogen) and oligo dT (Invitrogen) and random examers (Roche). After synthesis of cDNA (2–3 h at 42 °C), RNA is hydrolyzed by addition of sodium hydroxide and EDTA to a final concentration of 100 mM and 10 mM, respectively and incubated at 65 °C for 10 min. The hydrolysis reaction is neutralized with 1 M HEPES. After removing free nucleotides by purification and concentration using Microcon-30 microconcentrators, the aminoallyl-labeled samples are coupled to succinimidyl ester of cyanine-3 (Cy3) and cyanine-5 (Cy5) (Amersham) combined with 1 M NaHCO3, pH 9. Coupling takes place in the dark at 25 °C for 1 h. Appropriate Cy3 and Cy5 labeled cDNA samples are purified following Qiagen Qiaquick PCR Purification Kit instructions. Poly-dA (12–18 mer), 20× SSC, and HEPES pH 7.0 are added. After the resultant mix was filtered through a Millipore 0.45 ?M filter, 10% SDS is added. The samples are incubated for 2 min at 100 °C and cooled in a microcentrifuge prior to loading and then applied to microarray.
-
Hybridization
Incubation at 65 °C for 12–15 h.
-
Washing
Hybridized slides are washed in a solution of water, 20× SSC, and 10% SDS, rinsed in water and 20× SSC, and dried via centrifugation for 2 min at 1000 rpm.
-
Image acquisition
The arrays are scanned immediately. Each comparison is performed in duplicate. Fluorescent cDNA bound to the microarray is detected with a GenePix 4000 microarray scanner (Axon Instruments, Foster City, CA), using the GenePix 4000 software package to quantify microarray fluorescence.
-
Adjustment of intensity values
Intensity values are adjusted by subtracting surrounding background from spots. The median of spot intensities is corrected for background. To eliminate signals that are most prone to estimation error, any spot is excluded from analysis if both the Cy3 and Cy5 mean fluorescence signals are within two standard deviations of the mean background signals for that spot. This procedure avoids artificially inflated measurements of expression due to low signals. Additional to eliminating flagged spots, spots are also visually inspected and flawed ones discarded from the analysis.
-
Normalization
Data are normalized to mean ratio intensity.
- biomaterial type
- Saccharomyces cerevisiae
created over 17 years ago
(2 March 2009)
last modified over 14 years ago
(28 September 2011)
[ RDF  ]
[ RelFinder
]
[ RelFinder  ]
]
